Models without compartments ($PRED)
This vignette introduces a new formal code block for writing models where there are no compartments. The block is named after the analogous NONMEM block called $PRED. This functionality has always been possible with mrgsolve, but only now is there a code block dedicated to these models. Also, a relaxed set of data set constraints have been put in place when these types of models are invoked.
Status
The functionality in this vignette can only be access from the GitHub version. We will update this vignette once these features are rolled into a release on CRAN.
Example model
As a most-basic model, we look at the pred1 model in modlib()
The model code is
This is a random-intercept, random slope linear model. Like other models in mrgsolve, you can write parameters ($PARAM), and random effects ($OMEGA). But the model is actually written in $PRED.
When mrgsolve finds $PRED, it will generate an error if it also finds $MAIN, $TABLE, or $ODE. However, the code that gets entered into $PRED would function exactly as if you put it in $TABLE.
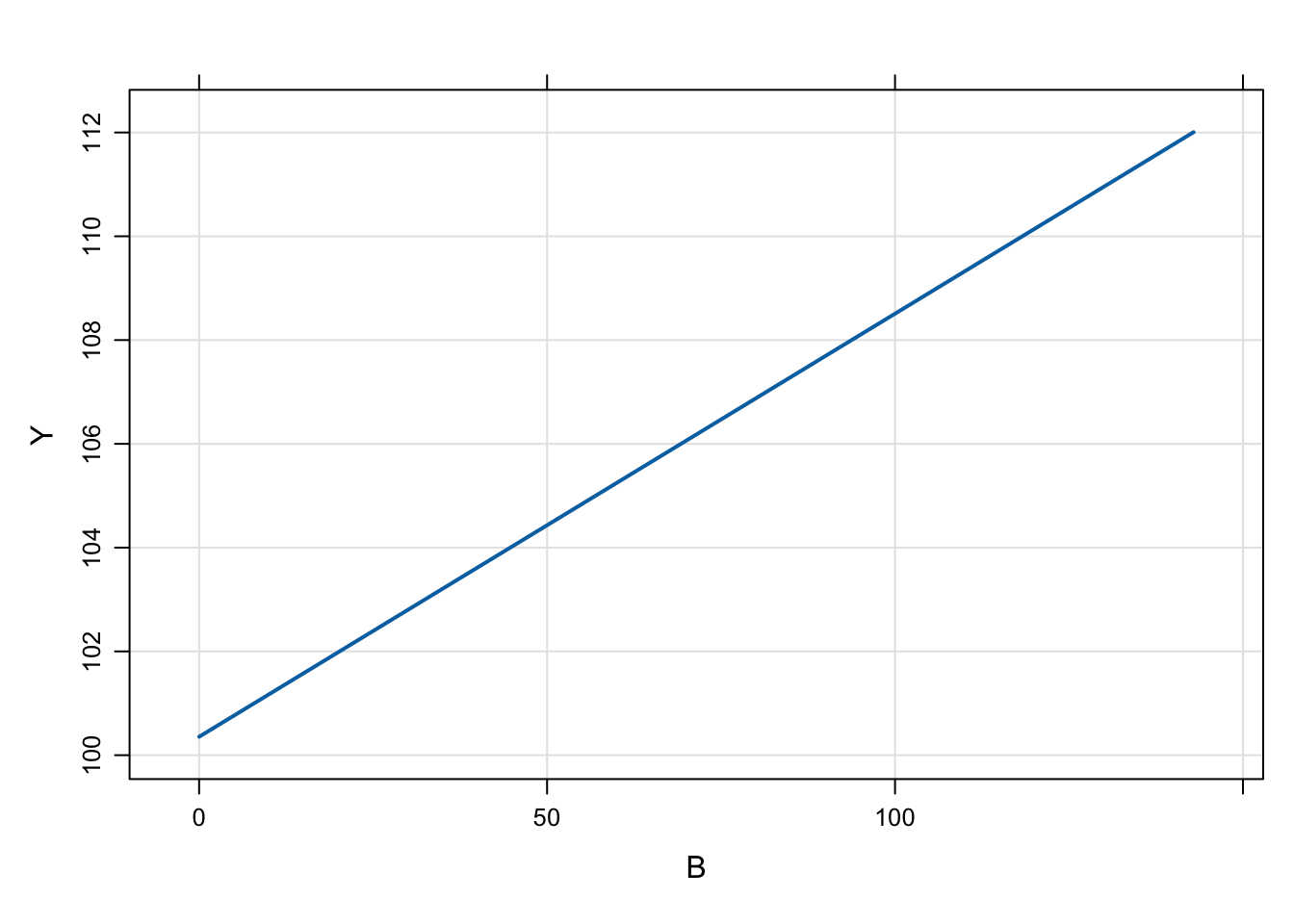
In the example model, the response is a function of the parameter B, so we’ll generate an input data set with some values of B
Warning: `data_frame()` was deprecated in tibble 1.1.0.
ℹ Please use `tibble()` instead.# A tibble: 6 × 2
ID B
<dbl> <dbl>
1 1 2.83
2 1 2.36
3 1 0.331
4 1 0.466
5 1 8.34
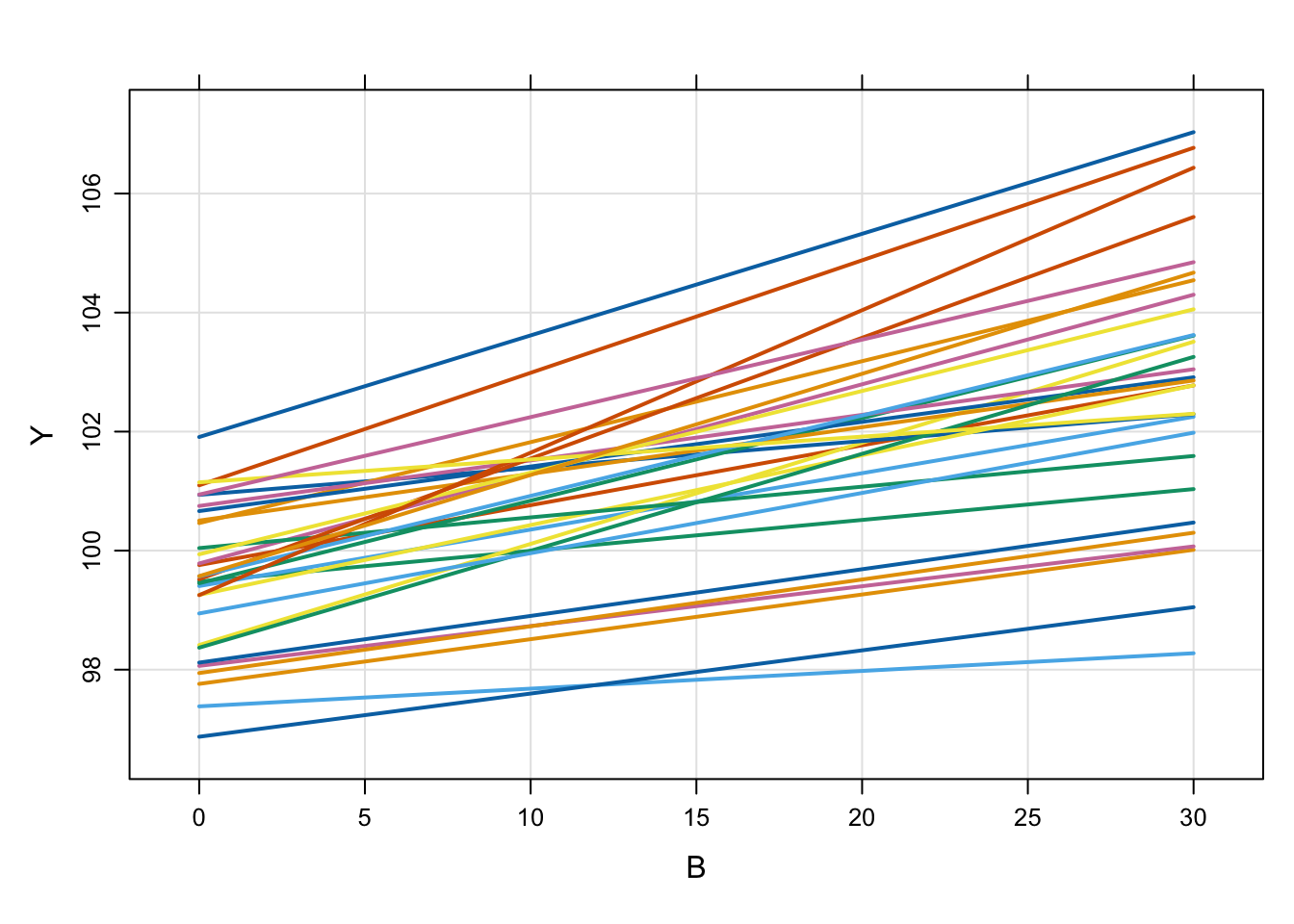
6 1 39.6 Like other models, we can simulate from a population
PK/PD Model
Here is an implementation of a PK/PD model using $PRED
In this model
- Calculate
CLas a function ofWTand a random effect - Derive
AUCfromCLandDOSE - The response (
Y) is a calculated fromAUCand the Emax model parameters
To simulate, look at 50 subjects at each of 5 doses
data <-
expand.idata(DOSE = c(30,50,80,110,200),ID = 1:50) %>%
mutate(WT = exp(rnorm(n(),log(80),1)))
head(data) ID DOSE WT
1 1 30 59.13254
2 2 50 317.32739
3 3 80 242.15746
4 4 110 170.78136
5 5 200 248.18054
6 6 30 51.22012 ID time WT DOSE AUC Y
1 1 0 59.13254 30 231.90852 110.61330
2 2 0 317.32739 50 36.76051 85.27834
3 3 0 242.15746 80 36.54808 98.90407
4 4 0 170.78136 110 23.68354 80.29131
5 5 0 248.18054 200 331.10229 108.81926
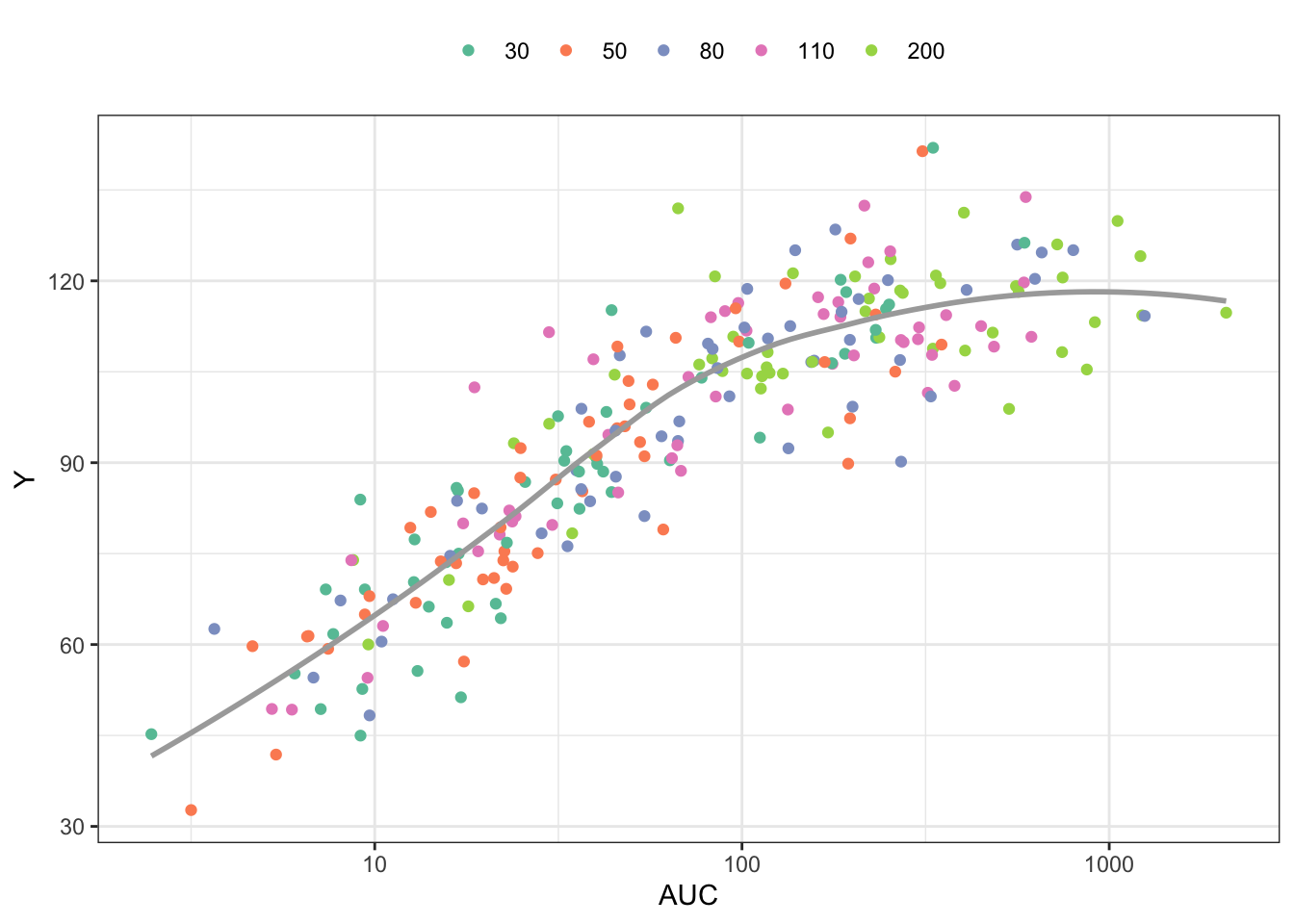
6 6 0 51.22012 30 251.58373 116.09649Plot the response (Y) versus AUC, colored by dose
library(ggplot2)
ggplot(out, aes(AUC,Y,col =factor(DOSE))) +
geom_point() +
scale_x_continuous(trans = "log", breaks = 10^seq(-4,4)) +
geom_smooth(aes(AUC,Y),se = FALSE,col="darkgrey") + theme_bw() +
scale_color_brewer(palette = "Set2", name = "") +
theme(legend.position = "top")`geom_smooth()` using method = 'loess' and formula = 'y ~ x'