library(ggplot2)
library(mrgsolve)
library(nloptr)
library(dplyr)
library(magrittr)
options(mrgsolve.soloc="build")This is a shortened version of map_bayes.html, showing only the estimation step.
1 The model
We code up a one-compartment model, with fixed TVCL and TVV. We have ETA1 and ETA2 as parameters in the model so that we can update them as the optimizer proceeds. Our goal is to find the values of ETA that are most consistent with the data.
code <- '
$SET request=""
$PARAM ETA1 = 0, ETA2 = 0
$CMT CENT
$PKMODEL ncmt=1
$MAIN
double TVCL = 1.5;
double TVVC = 23.4;
double CL = TVCL*exp(ETA1);
double V = TVVC*exp(ETA2);
$TABLE
capture DV = (CENT/V);
'
mod <- mcode_cache("map", code)We also have the OMEGA and SIGMA matrices from the population model estimation step.
omega <- cmat(0.23,-0.78, 0.62)
omega.inv <- solve(omega)
sigma <- matrix(0.0032)Read in the data set. Notice that DV has value NA for dosing records. When calculating the joint likelihood of all the data, we will remove the missing values (dosing records don’t have observations that contribute to the likelihood value).
data <- readRDS("map_bayes_data.RDS")
head(data)# A tibble: 4 × 6
ID time evid amt cmt DV
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 0 1 750 1 NA
2 1 0.5 0 0 0 42.7
3 1 12 0 0 0 6.93
4 1 24 0 0 0 1.372 Optimize
This function takes in a set of proposed \(\eta\)s along with the observed data vector, the data set and a model object and returns the value of the EBE objective function
When we do the estimation, the fixed effects and random effect variances are fixed.
The estimates are the \(\eta\) for clearance and volume
Arguments:
etathe current values from the optimizerycolthe observed data column namedthe data setmthe model objectdvcolthe predicted data column namepredifTRUE, just return predicted values
2.1 What is this function doing?
- get the matrix for residual error
- Make sure
etais a list - Make sure
etais properly named (i.e.ETA1andETA2) - Copy
etainto a matrix that is one row - Update the model object (
m) with the current values ofETA1andETA2 - Simulate from data set
dand save output tooutobject - If we are just requesting predictions (
if(pred)) return the simulated data - The final lines calculate the EBE objective function; see this paper for reference
- Notice that the function returns a single value (a number); the optimizer will minimize this value
mapbayes <- function(eta,d,ycol,m,dvcol=ycol,pred=FALSE) {
sig2 <- as.numeric(sigma)
eta <- as.list(eta)
names(eta) <- names(init)
eta_m <- eta %>% unlist %>% matrix(nrow=1)
m <- param(m,eta)
out <- mrgsim(m,data=d,output="df")
if(pred) return(out)
# http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3339294/
sig2j <- out[[dvcol]]^2*sig2
sqwres <- log(sig2j) + (1/sig2j)*(d[[ycol]] - out[[dvcol]])^2
nOn <- diag(eta_m %*% omega.inv %*% t(eta_m))
return(sum(sqwres,na.rm=TRUE) + nOn)
}2.2 Initial estimate
- Note again that we are optimizing the etas here
init <- c(ETA1=-0.3, ETA2=0.2)Fit the data
newuoais from thenloptrpackage- Other optimizers (via
optim) could probably also be used
Arguments to newuoa
- First: the initial estimates
- Second: the function to optimize
- The other argument are passed to
mapbayes
fit <- nloptr::newuoa(init,mapbayes,ycol="DV",m=mod,d=data)Here are the final estimates
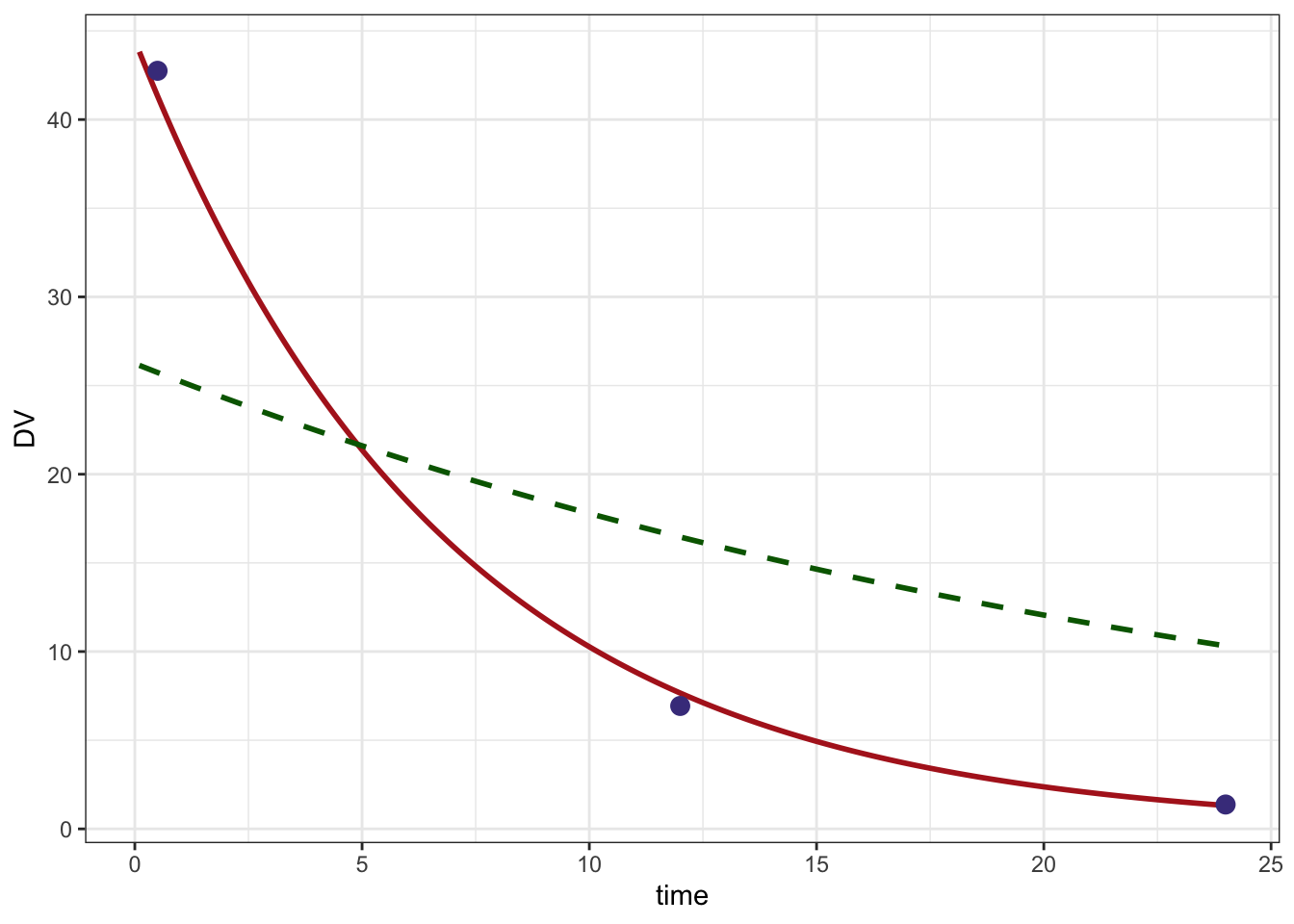
fit$par[1] 0.4995400 -0.32748583 Look at the result
A data set and model to get predictions; this will give us a smooth prediction line
pdata <- data %>% filter(evid==1)
pmod <- mod %>% update(end=24, delta=0.1) Predicted line based on final estimates
pred <- mapbayes(fit$par,ycol="DV",pdata,pmod,pred=TRUE) %>% filter(time > 0)
head(pred) ID time DV
1 1 0.1 43.82331
2 1 0.2 43.18567
3 1 0.3 42.55731
4 1 0.4 41.93809
5 1 0.5 41.32789
6 1 0.6 40.72656Predicted line based on initial estimates
initial <- mapbayes(init,ycol="DV",pdata,pmod,,pred=TRUE) %>% filter(time > 0)
head(initial) ID time DV
1 1 0.1 26.13954
2 1 0.2 26.03811
3 1 0.3 25.93707
4 1 0.4 25.83642
5 1 0.5 25.73616
6 1 0.6 25.63629Plot
ggplot() +
geom_line(data=pred, aes(time,DV),col="firebrick", lwd=1) +
geom_line(data=initial,aes(time,DV), lty=2, col="darkgreen", lwd=1) +
geom_point(data=data %>% filter(evid==0), aes(time,DV), col="darkslateblue",size=3) +
theme_bw()